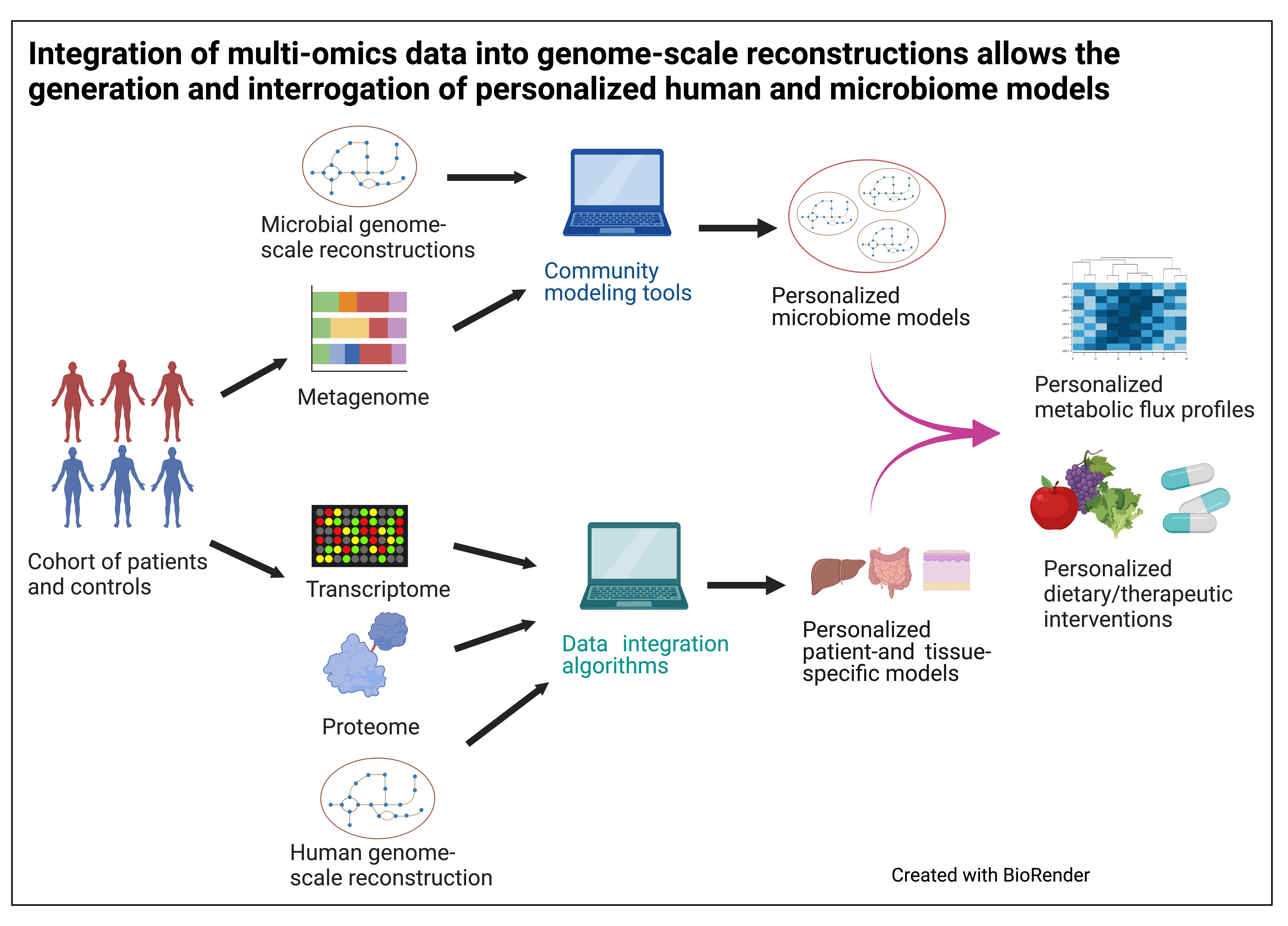
About the courseThe development of high-throughput omics methods such as genomics, transcriptomics, proteomics, and metagenomics has led to an avalanche of available data. This has motivated the field of systems biology, which aims to comprehensively characterize the components of biological systems such as cells, organisms, or populations to understand their function and interpret omics data in their biological context. One powerful systems biology approach is Constraint-Based Reconstruction and Analysis (COBRA). The COBRA method relies on manually refined genome-scale reconstructions that can be converted into mathematical models and interrogated through simulations. Integrating transcriptomic, proteomic, or metagenomic data into genome-scale models enables the generation of personalized models that can stratify samples by the computed metabolic fluxes. Personalized modeling could result in novel insight into disease mechanisms and has valuable applications in precision medicine, such as designing targeted treatment. In this course, state of the art methods for the integration of omics data into genome-scale models will be taught. One focus of the course will be the use of transcriptomic and proteomic data to construct tissue-and context-specific models. Applications of context-specific models for the analysis of disease states and in precision medicine will be reviewed. Hands-on tutorials will enable participants to apply algorithms for the generation of context-specific models to their data. Another focus will be the generation of multi-species models of microbiome communities from metagenomics data. Recent applications of microbiome community modeling to elucidate host-microbiome interactions in diseases will be reviewed. Tutorials will give participants hands-on experience in integrating metagenomics data into microbiome community models and predicting community-level metabolism. The course will end with a round table where participants can discuss strategies and challenges in their research.
Specific learning outcomes
|